| Locus: |
|
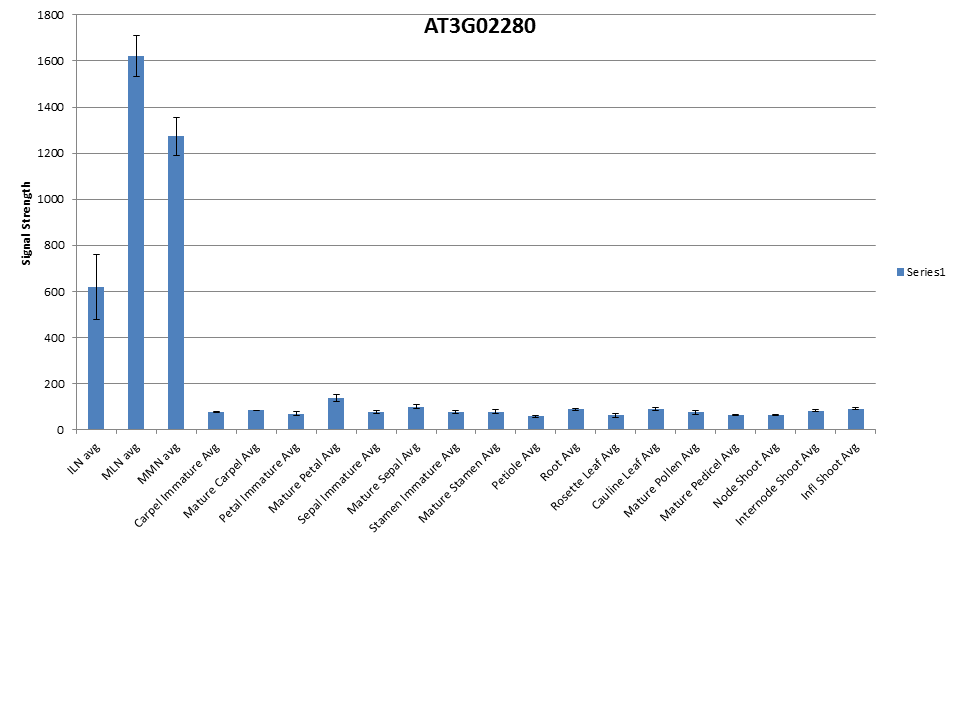
AT3G02280 |
| Name: |
|
|
| Description: |
|
Flavodoxin family protein; FUNCTIONS IN: oxidoreductase activity, FMN binding; INVOLVED IN: oxidation reduction; LOCATED IN: cellular_component unknown; EXPRESSED IN: 18 plant structures; EXPRESSED DURING: 7 growth stages; CONTAINS InterPro DOMAIN/s: Oxidoreductase FAD/NAD(P)-binding (InterPro:IPR001433), Ferredoxin reductase-type FAD-binding domain (InterPro:IPR017927), Riboflavin synthase-like beta-barrel (InterPro:IPR017938), FAD-binding, type 1 (InterPro:IPR003097), Flavodoxin/nitric oxide synthase (InterPro:IPR008254), Flavoprotein pyridine nucleotide cytochrome reductase (InterPro:IPR001709); BEST Arabidopsis thaliana protein match is: P450 reductase 1 (TAIR:AT4G24520.1); Has 7053 Blast hits to 6598 proteins in 1587 species: Archae - 9; Bacteria - 3832; Metazoa - 1042; Fungi - 865; Plants - 492; Viruses - 0; Other Eukaryotes - 813 (source: NCBI BLink). |
| Associations: |
|
| Category |
|
Relationship Type |
|
Keyword |
| GO Biological Process |
|
involved in |
|
oxidation-reduction process
|
| GO Cellular Component |
|
located in |
|
cellular_component
|
| GO Molecular Function |
|
functions in |
|
FMN binding
|
| GO Molecular Function |
|
has |
|
oxidoreductase activity
|
| Growth and Developmental Stages |
|
expressed during |
|
4 anthesis ,C globular stage ,D bilateral stage ,E expanded cotyledon stage ,F mature embryo stage ,LP.04 four leaves visible ,petal differentiation and expansion stage
|
| Plant structure |
|
expressed in |
|
carpel ,collective leaf structure ,flower ,hypocotyl ,inflorescence meristem ,leaf lamina base ,pedicel ,petal ,plant embryo ,pollen ,root ,seed ,sepal ,shoot apex ,shoot system ,stamen ,stem ,vascular leaf
|
|
|
Mutants: |
|
CS822043 (line202) SALK_123688 (line203) |
| wild type tissue expression: |
|
| wild type tissue expression ... |
|
|
|